Hidden Markov model
A hidden Markov model (HMM) is a Markov model in which the observations are dependent on a latent (or hidden) Markov process (referred to as ). An HMM requires that there be an observable process whose outcomes depend on the outcomes of in a known way. Since cannot be observed directly, the goal is to learn about state of by observing . By definition of being a Markov model, an HMM has an additional requirement that the outcome of at time must be "influenced" exclusively by the outcome of at and that the outcomes of and at must be conditionally independent of at given at time . Estimation of the parameters in an HMM can be performed using maximum likelihood estimation. For linear chain HMMs, the Baum–Welch algorithm can be used to estimate parameters.
Hidden Markov models are known for their applications to thermodynamics, statistical mechanics, physics, chemistry, economics, finance, signal processing, information theory, pattern recognition—such as speech,[1] handwriting, gesture recognition,[2] part-of-speech tagging, musical score following,[3] partial discharges[4] and bioinformatics.[5][6]
Definition
[edit]Let and be discrete-time stochastic processes and . The pair is a hidden Markov model if
- is a Markov process whose behavior is not directly observable ("hidden");
- ,
- for every , , and every Borel set .
Let and be continuous-time stochastic processes. The pair is a hidden Markov model if
- is a Markov process whose behavior is not directly observable ("hidden");
- ,
- for every , every Borel set , and every family of Borel sets .
Terminology
[edit]The states of the process (resp. are called hidden states, and (resp. is called emission probability or output probability.
Examples
[edit]Drawing balls from hidden urns
[edit]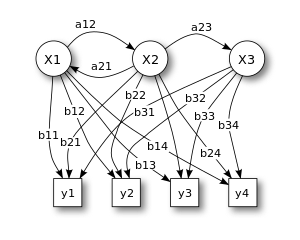
X — states
y — possible observations
a — state transition probabilities
b — output probabilities
In its discrete form, a hidden Markov process can be visualized as a generalization of the urn problem with replacement (where each item from the urn is returned to the original urn before the next step).[7] Consider this example: in a room that is not visible to an observer there is a genie. The room contains urns X1, X2, X3, ... each of which contains a known mix of balls, with each ball having a unique label y1, y2, y3, ... . The genie chooses an urn in that room and randomly draws a ball from that urn. It then puts the ball onto a conveyor belt, where the observer can observe the sequence of the balls but not the sequence of urns from which they were drawn. The genie has some procedure to choose urns; the choice of the urn for the n-th ball depends only upon a random number and the choice of the urn for the (n − 1)-th ball. The choice of urn does not directly depend on the urns chosen before this single previous urn; therefore, this is called a Markov process. It can be described by the upper part of Figure 1.
The Markov process cannot be observed, only the sequence of labeled balls, thus this arrangement is called a hidden Markov process. This is illustrated by the lower part of the diagram shown in Figure 1, where one can see that balls y1, y2, y3, y4 can be drawn at each state. Even if the observer knows the composition of the urns and has just observed a sequence of three balls, e.g. y1, y2 and y3 on the conveyor belt, the observer still cannot be sure which urn (i.e., at which state) the genie has drawn the third ball from. However, the observer can work out other information, such as the likelihood that the third ball came from each of the urns.
Weather guessing game
[edit]Consider two friends, Alice and Bob, who live far apart from each other and who talk together daily over the telephone about what they did that day. Bob is only interested in three activities: walking in the park, shopping, and cleaning his apartment. The choice of what to do is determined exclusively by the weather on a given day. Alice has no definite information about the weather, but she knows general trends. Based on what Bob tells her he did each day, Alice tries to guess what the weather must have been like.
Alice believes that the weather operates as a discrete Markov chain. There are two states, "Rainy" and "Sunny", but she cannot observe them directly, that is, they are hidden from her. On each day, there is a certain chance that Bob will perform one of the following activities, depending on the weather: "walk", "shop", or "clean". Since Bob tells Alice about his activities, those are the observations. The entire system is that of a hidden Markov model (HMM).
Alice knows the general weather trends in the area, and what Bob likes to do on average. In other words, the parameters of the HMM are known. They can be represented as follows in Python:
states = ("Rainy", "Sunny")
observations = ("walk", "shop", "clean")
start_probability = {"Rainy": 0.6, "Sunny": 0.4}
transition_probability = {
"Rainy": {"Rainy": 0.7, "Sunny": 0.3},
"Sunny": {"Rainy": 0.4, "Sunny": 0.6},
}
emission_probability = {
"Rainy": {"walk": 0.1, "shop": 0.4, "clean": 0.5},
"Sunny": {"walk": 0.6, "shop": 0.3, "clean": 0.1},
}
In this piece of code, start_probability represents Alice's belief about which state the HMM is in when Bob first calls her (all she knows is that it tends to be rainy on average). The particular probability distribution used here is not the equilibrium one, which is (given the transition probabilities) approximately {'Rainy': 0.57, 'Sunny': 0.43}. The transition_probability represents the change of the weather in the underlying Markov chain. In this example, there is only a 30% chance that tomorrow will be sunny if today is rainy. The emission_probability represents how likely Bob is to perform a certain activity on each day. If it is rainy, there is a 50% chance that he is cleaning his apartment; if it is sunny, there is a 60% chance that he is outside for a walk.

A similar example is further elaborated in the Viterbi algorithm page.
Structural architecture
[edit]The diagram below shows the general architecture of an instantiated HMM. Each oval shape represents a random variable that can adopt any of a number of values. The random variable x(t) is the hidden state at time t (with the model from the above diagram, x(t) ∈ { x1, x2, x3 }). The random variable y(t) is the observation at time t (with y(t) ∈ { y1, y2, y3, y4 }). The arrows in the diagram (often called a trellis diagram) denote conditional dependencies.
From the diagram, it is clear that the conditional probability distribution of the hidden variable x(t) at time t, given the values of the hidden variable x at all times, depends only on the value of the hidden variable x(t − 1); the values at time t − 2 and before have no influence. This is called the Markov property. Similarly, the value of the observed variable y(t) depends on only the value of the hidden variable x(t) (both at time t).
In the standard type of hidden Markov model considered here, the state space of the hidden variables is discrete, while the observations themselves can either be discrete (typically generated from a categorical distribution) or continuous (typically from a Gaussian distribution). The parameters of a hidden Markov model are of two types, transition probabilities and emission probabilities (also known as output probabilities). The transition probabilities control the way the hidden state at time t is chosen given the hidden state at time .
The hidden state space is assumed to consist of one of N possible values, modelled as a categorical distribution. (See the section below on extensions for other possibilities.) This means that for each of the N possible states that a hidden variable at time t can be in, there is a transition probability from this state to each of the N possible states of the hidden variable at time , for a total of transition probabilities. The set of transition probabilities for transitions from any given state must sum to 1. Thus, the matrix of transition probabilities is a Markov matrix. Because any transition probability can be determined once the others are known, there are a total of transition parameters.
In addition, for each of the N possible states, there is a set of emission probabilities governing the distribution of the observed variable at a particular time given the state of the hidden variable at that time. The size of this set depends on the nature of the observed variable. For example, if the observed variable is discrete with M possible values, governed by a categorical distribution, there will be separate parameters, for a total of emission parameters over all hidden states. On the other hand, if the observed variable is an M-dimensional vector distributed according to an arbitrary multivariate Gaussian distribution, there will be M parameters controlling the means and parameters controlling the covariance matrix, for a total of emission parameters. (In such a case, unless the value of M is small, it may be more practical to restrict the nature of the covariances between individual elements of the observation vector, e.g. by assuming that the elements are independent of each other, or less restrictively, are independent of all but a fixed number of adjacent elements.)

Inference
[edit]
5 3 2 5 3 2
4 3 2 5 3 2
3 1 2 5 3 2
The most likely sequence can be found by evaluating the joint probability of both the state sequence and the observations for each case (simply by multiplying the probability values, which here correspond to the opacities of the arrows involved). In general, this type of problem (i.e., finding the most likely explanation for an observation sequence) can be solved efficiently using the Viterbi algorithm.
Several inference problems are associated with hidden Markov models, as outlined below.
Probability of an observed sequence
[edit]The task is to compute in a best way, given the parameters of the model, the probability of a particular output sequence. This requires summation over all possible state sequences:
The probability of observing a sequence
- ,
of length L is given by
- ,
where the sum runs over all possible hidden-node sequences
- .
Applying the principle of dynamic programming, this problem, too, can be handled efficiently using the forward algorithm.
Probability of the latent variables
[edit]A number of related tasks ask about the probability of one or more of the latent variables, given the model's parameters and a sequence of observations .
Filtering
[edit]The task is to compute, given the model's parameters and a sequence of observations, the distribution over hidden states of the last latent variable at the end of the sequence, i.e. to compute . This task is used when the sequence of latent variables is thought of as the underlying states that a process moves through at a sequence of points in time, with corresponding observations at each point. Then, it is natural to ask about the state of the process at the end.
This problem can be handled efficiently using the forward algorithm. An example is when the algorithm is applied to a Hidden Markov Network to determine .
Smoothing
[edit]This is similar to filtering but asks about the distribution of a latent variable somewhere in the middle of a sequence, i.e. to compute for some . From the perspective described above, this can be thought of as the probability distribution over hidden states for a point in time k in the past, relative to time t.
The forward-backward algorithm is a good method for computing the smoothed values for all hidden state variables.
Most likely explanation
[edit]The task, unlike the previous two, asks about the joint probability of the entire sequence of hidden states that generated a particular sequence of observations (see illustration on the right). This task is generally applicable when HMM's are applied to different sorts of problems from those for which the tasks of filtering and smoothing are applicable. An example is part-of-speech tagging, where the hidden states represent the underlying parts of speech corresponding to an observed sequence of words. In this case, what is of interest is the entire sequence of parts of speech, rather than simply the part of speech for a single word, as filtering or smoothing would compute.
This task requires finding a maximum over all possible state sequences, and can be solved efficiently by the Viterbi algorithm.
Statistical significance
[edit]For some of the above problems, it may also be interesting to ask about statistical significance. What is the probability that a sequence drawn from some null distribution will have an HMM probability (in the case of the forward algorithm) or a maximum state sequence probability (in the case of the Viterbi algorithm) at least as large as that of a particular output sequence?[8] When an HMM is used to evaluate the relevance of a hypothesis for a particular output sequence, the statistical significance indicates the false positive rate associated with failing to reject the hypothesis for the output sequence.
Learning
[edit]The parameter learning task in HMMs is to find, given an output sequence or a set of such sequences, the best set of state transition and emission probabilities. The task is usually to derive the maximum likelihood estimate of the parameters of the HMM given the set of output sequences. No tractable algorithm is known for solving this problem exactly, but a local maximum likelihood can be derived efficiently using the Baum–Welch algorithm or the Baldi–Chauvin algorithm. The Baum–Welch algorithm is a special case of the expectation-maximization algorithm.
If the HMMs are used for time series prediction, more sophisticated Bayesian inference methods, like Markov chain Monte Carlo (MCMC) sampling are proven to be favorable over finding a single maximum likelihood model both in terms of accuracy and stability.[9] Since MCMC imposes significant computational burden, in cases where computational scalability is also of interest, one may alternatively resort to variational approximations to Bayesian inference, e.g.[10] Indeed, approximate variational inference offers computational efficiency comparable to expectation-maximization, while yielding an accuracy profile only slightly inferior to exact MCMC-type Bayesian inference.
Applications
[edit]
HMMs can be applied in many fields where the goal is to recover a data sequence that is not immediately observable (but other data that depend on the sequence are). Applications include:
- Computational finance[11][12]
- Single-molecule kinetic analysis[13]
- Neuroscience[14][15]
- Cryptanalysis
- Speech recognition, including Siri[16]
- Speech synthesis
- Part-of-speech tagging
- Document separation in scanning solutions
- Machine translation
- Partial discharge
- Gene prediction
- Handwriting recognition[17]
- Alignment of bio-sequences
- Time series analysis
- Activity recognition
- Protein folding[18]
- Sequence classification[19]
- Metamorphic virus detection[20]
- Sequence motif discovery (DNA and proteins)[21]
- DNA hybridization kinetics[22][23]
- Chromatin state discovery[24]
- Transportation forecasting[25]
- Solar irradiance variability[26][27][28]
History
[edit]Hidden Markov models were described in a series of statistical papers by Leonard E. Baum and other authors in the second half of the 1960s.[29][30][31][32][33] One of the first applications of HMMs was speech recognition, starting in the mid-1970s.[34][35][36][37] From the linguistics point of view, hidden Markov models are equivalent to stochastic regular grammar.[38]
In the second half of the 1980s, HMMs began to be applied to the analysis of biological sequences,[39] in particular DNA. Since then, they have become ubiquitous in the field of bioinformatics.[40]
Extensions
[edit]General state spaces
[edit]In the hidden Markov models considered above, the state space of the hidden variables is discrete, while the observations themselves can either be discrete (typically generated from a categorical distribution) or continuous (typically from a Gaussian distribution). Hidden Markov models can also be generalized to allow continuous state spaces. Examples of such models are those where the Markov process over hidden variables is a linear dynamical system, with a linear relationship among related variables and where all hidden and observed variables follow a Gaussian distribution. In simple cases, such as the linear dynamical system just mentioned, exact inference is tractable (in this case, using the Kalman filter); however, in general, exact inference in HMMs with continuous latent variables is infeasible, and approximate methods must be used, such as the extended Kalman filter or the particle filter.
Nowadays, inference in hidden Markov models is performed in nonparametric settings, where the dependency structure enables identifiability of the model[41] and the learnability limits are still under exploration.[42]
Bayesian modeling of the transitions probabilities
[edit]Hidden Markov models are generative models, in which the joint distribution of observations and hidden states, or equivalently both the prior distribution of hidden states (the transition probabilities) and conditional distribution of observations given states (the emission probabilities), is modeled. The above algorithms implicitly assume a uniform prior distribution over the transition probabilities. However, it is also possible to create hidden Markov models with other types of prior distributions. An obvious candidate, given the categorical distribution of the transition probabilities, is the Dirichlet distribution, which is the conjugate prior distribution of the categorical distribution. Typically, a symmetric Dirichlet distribution is chosen, reflecting ignorance about which states are inherently more likely than others. The single parameter of this distribution (termed the concentration parameter) controls the relative density or sparseness of the resulting transition matrix. A choice of 1 yields a uniform distribution. Values greater than 1 produce a dense matrix, in which the transition probabilities between pairs of states are likely to be nearly equal. Values less than 1 result in a sparse matrix in which, for each given source state, only a small number of destination states have non-negligible transition probabilities. It is also possible to use a two-level prior Dirichlet distribution, in which one Dirichlet distribution (the upper distribution) governs the parameters of another Dirichlet distribution (the lower distribution), which in turn governs the transition probabilities. The upper distribution governs the overall distribution of states, determining how likely each state is to occur; its concentration parameter determines the density or sparseness of states. Such a two-level prior distribution, where both concentration parameters are set to produce sparse distributions, might be useful for example in unsupervised part-of-speech tagging, where some parts of speech occur much more commonly than others; learning algorithms that assume a uniform prior distribution generally perform poorly on this task. The parameters of models of this sort, with non-uniform prior distributions, can be learned using Gibbs sampling or extended versions of the expectation-maximization algorithm.
An extension of the previously described hidden Markov models with Dirichlet priors uses a Dirichlet process in place of a Dirichlet distribution. This type of model allows for an unknown and potentially infinite number of states. It is common to use a two-level Dirichlet process, similar to the previously described model with two levels of Dirichlet distributions. Such a model is called a hierarchical Dirichlet process hidden Markov model, or HDP-HMM for short. It was originally described under the name "Infinite Hidden Markov Model"[43] and was further formalized in "Hierarchical Dirichlet Processes".[44]
Discriminative approach
[edit]A different type of extension uses a discriminative model in place of the generative model of standard HMMs. This type of model directly models the conditional distribution of the hidden states given the observations, rather than modeling the joint distribution. An example of this model is the so-called maximum entropy Markov model (MEMM), which models the conditional distribution of the states using logistic regression (also known as a "maximum entropy model"). The advantage of this type of model is that arbitrary features (i.e. functions) of the observations can be modeled, allowing domain-specific knowledge of the problem at hand to be injected into the model. Models of this sort are not limited to modeling direct dependencies between a hidden state and its associated observation; rather, features of nearby observations, of combinations of the associated observation and nearby observations, or in fact of arbitrary observations at any distance from a given hidden state can be included in the process used to determine the value of a hidden state. Furthermore, there is no need for these features to be statistically independent of each other, as would be the case if such features were used in a generative model. Finally, arbitrary features over pairs of adjacent hidden states can be used rather than simple transition probabilities. The disadvantages of such models are: (1) The types of prior distributions that can be placed on hidden states are severely limited; (2) It is not possible to predict the probability of seeing an arbitrary observation. This second limitation is often not an issue in practice, since many common usages of HMM's do not require such predictive probabilities.
A variant of the previously described discriminative model is the linear-chain conditional random field. This uses an undirected graphical model (aka Markov random field) rather than the directed graphical models of MEMM's and similar models. The advantage of this type of model is that it does not suffer from the so-called label bias problem of MEMM's, and thus may make more accurate predictions. The disadvantage is that training can be slower than for MEMM's.
Other extensions
[edit]Yet another variant is the factorial hidden Markov model, which allows for a single observation to be conditioned on the corresponding hidden variables of a set of independent Markov chains, rather than a single Markov chain. It is equivalent to a single HMM, with states (assuming there are states for each chain), and therefore, learning in such a model is difficult: for a sequence of length , a straightforward Viterbi algorithm has complexity . To find an exact solution, a junction tree algorithm could be used, but it results in an complexity. In practice, approximate techniques, such as variational approaches, could be used.[45]
All of the above models can be extended to allow for more distant dependencies among hidden states, e.g. allowing for a given state to be dependent on the previous two or three states rather than a single previous state; i.e. the transition probabilities are extended to encompass sets of three or four adjacent states (or in general adjacent states). The disadvantage of such models is that dynamic-programming algorithms for training them have an running time, for adjacent states and total observations (i.e. a length- Markov chain). This extension has been widely used in bioinformatics, in the modeling of DNA sequences.
Another recent extension is the triplet Markov model,[46] in which an auxiliary underlying process is added to model some data specificities. Many variants of this model have been proposed. One should also mention the interesting link that has been established between the theory of evidence and the triplet Markov models[47] and which allows to fuse data in Markovian context[48] and to model nonstationary data.[49][50] Alternative multi-stream data fusion strategies have also been proposed in recent literature, e.g.,[51]
Finally, a different rationale towards addressing the problem of modeling nonstationary data by means of hidden Markov models was suggested in 2012.[52] It consists in employing a small recurrent neural network (RNN), specifically a reservoir network,[53] to capture the evolution of the temporal dynamics in the observed data. This information, encoded in the form of a high-dimensional vector, is used as a conditioning variable of the HMM state transition probabilities. Under such a setup, eventually is obtained a nonstationary HMM, the transition probabilities of which evolve over time in a manner that is inferred from the data, in contrast to some unrealistic ad-hoc model of temporal evolution.
In 2023, two innovative algorithms were introduced for the Hidden Markov Model. These algorithms enable the computation of the posterior distribution of the HMM without the necessity of explicitly modeling the joint distribution, utilizing only the conditional distributions.[54][55] Unlike traditional methods such as the Forward-Backward and Viterbi algorithms, which require knowledge of the joint law of the HMM and can be computationally intensive to learn, the Discriminative Forward-Backward and Discriminative Viterbi algorithms circumvent the need for the observation's law.[56][57] This breakthrough allows the HMM to be applied as a discriminative model, offering a more efficient and versatile approach to leveraging Hidden Markov Models in various applications.
The model suitable in the context of longitudinal data is named latent Markov model.[58] The basic version of this model has been extended to include individual covariates, random effects and to model more complex data structures such as multilevel data. A complete overview of the latent Markov models, with special attention to the model assumptions and to their practical use is provided in[59]
Measure theory
[edit]
Given a Markov transition matrix and an invariant distribution on the states, a probability measure can be imposed on the set of subshifts. For example, consider the Markov chain given on the left on the states , with invariant distribution . By ignoring the distinction between , this space of subshifts is projected on into another space of subshifts on , and this projection also projects the probability measure down to a probability measure on the subshifts on .
The curious thing is that the probability measure on the subshifts on is not created by a Markov chain on , not even multiple orders. Intuitively, this is because if one observes a long sequence of , then one would become increasingly sure that the , meaning that the observable part of the system can be affected by something infinitely in the past.[60][61]
Conversely, there exists a space of subshifts on 6 symbols, projected to subshifts on 2 symbols, such that any Markov measure on the smaller subshift has a preimage measure that is not Markov of any order (example 2.6[61]).
See also
[edit]- Andrey Markov
- Baum–Welch algorithm
- Bayesian inference
- Bayesian programming
- Richard James Boys
- Conditional random field
- Estimation theory
- HH-suite (HHpred, HHsearch) free server and software for protein sequence searching
- HMMER, a free hidden Markov model program for protein sequence analysis
- Hidden Bernoulli model
- Hidden semi-Markov model
- Hierarchical hidden Markov model
- Layered hidden Markov model
- Sequential dynamical system
- Stochastic context-free grammar
- Time Series Analysis
- Variable-order Markov model
- Viterbi algorithm
References
[edit]- ^ "Google Scholar".
- ^ Thad Starner, Alex Pentland. Real-Time American Sign Language Visual Recognition From Video Using Hidden Markov Models. Master's Thesis, MIT, Feb 1995, Program in Media Arts
- ^ B. Pardo and W. Birmingham. Modeling Form for On-line Following of Musical Performances Archived 2012-02-06 at the Wayback Machine. AAAI-05 Proc., July 2005.
- ^ Satish L, Gururaj BI (April 2003). "Use of hidden Markov models for partial discharge pattern classification". IEEE Transactions on Dielectrics and Electrical Insulation.
- ^ Li, N; Stephens, M (December 2003). "Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data". Genetics. 165 (4): 2213–33. doi:10.1093/genetics/165.4.2213. PMC 1462870. PMID 14704198.
- ^ Ernst, Jason; Kellis, Manolis (March 2012). "ChromHMM: automating chromatin-state discovery and characterization". Nature Methods. 9 (3): 215–216. doi:10.1038/nmeth.1906. PMC 3577932. PMID 22373907.
- ^ Lawrence R. Rabiner (February 1989). "A tutorial on Hidden Markov Models and selected applications in speech recognition" (PDF). Proceedings of the IEEE. 77 (2): 257–286. CiteSeerX 10.1.1.381.3454. doi:10.1109/5.18626. S2CID 13618539. [1]
- ^ Newberg, L. (2009). "Error statistics of hidden Markov model and hidden Boltzmann model results". BMC Bioinformatics. 10: 212. doi:10.1186/1471-2105-10-212. PMC 2722652. PMID 19589158.

- ^ Sipos, I. Róbert. Parallel stratified MCMC sampling of AR-HMMs for stochastic time series prediction. In: Proceedings, 4th Stochastic Modeling Techniques and Data Analysis International Conference with Demographics Workshop (SMTDA2016), pp. 295-306. Valletta, 2016. PDF
- ^ Chatzis, Sotirios P.; Kosmopoulos, Dimitrios I. (2011). "A variational Bayesian methodology for hidden Markov models utilizing Student's-t mixtures" (PDF). Pattern Recognition. 44 (2): 295–306. Bibcode:2011PatRe..44..295C. CiteSeerX 10.1.1.629.6275. doi:10.1016/j.patcog.2010.09.001. Archived from the original (PDF) on 2011-04-01. Retrieved 2018-03-11.
- ^ Sipos, I. Róbert; Ceffer, Attila; Levendovszky, János (2016). "Parallel Optimization of Sparse Portfolios with AR-HMMs". Computational Economics. 49 (4): 563–578. doi:10.1007/s10614-016-9579-y. S2CID 61882456.
- ^ Petropoulos, Anastasios; Chatzis, Sotirios P.; Xanthopoulos, Stylianos (2016). "A novel corporate credit rating system based on Student's-t hidden Markov models". Expert Systems with Applications. 53: 87–105. doi:10.1016/j.eswa.2016.01.015.
- ^ Nicolai, Christopher (2013). "Solving Ion Channel Kinetics with the QuB Software". Biophysical Reviews and Letters. 8 (3n04): 191–211. doi:10.1142/S1793048013300053.
- ^ Higgins, Cameron; Vidaurre, Diego; Kolling, Nils; Liu, Yunzhe; Behrens, Tim; Woolrich, Mark (2022). "Spatiotemporally Resolved Multivariate Pattern Analysis for M/EEG". Human Brain Mapping. 43 (10): 3062–3085. doi:10.1002/hbm.25835. PMC 9188977. PMID 35302683.
- ^ Diomedi, S.; Vaccari, F. E.; Galletti, C.; Hadjidimitrakis, K.; Fattori, P. (2021-10-01). "Motor-like neural dynamics in two parietal areas during arm reaching". Progress in Neurobiology. 205: 102116. doi:10.1016/j.pneurobio.2021.102116. hdl:11585/834094. ISSN 0301-0082. PMID 34217822. S2CID 235703641.
- ^ Domingos, Pedro (2015). The Master Algorithm: How the Quest for the Ultimate Learning Machine Will Remake Our World. Basic Books. p. 37. ISBN 9780465061921.
- ^ Kundu, Amlan, Yang He, and Paramvir Bahl. "Recognition of handwritten word: first and second order hidden Markov model based approach[dead link]." Pattern recognition 22.3 (1989): 283-297.
- ^ Stigler, J.; Ziegler, F.; Gieseke, A.; Gebhardt, J. C. M.; Rief, M. (2011). "The Complex Folding Network of Single Calmodulin Molecules". Science. 334 (6055): 512–516. Bibcode:2011Sci...334..512S. doi:10.1126/science.1207598. PMID 22034433. S2CID 5502662.
- ^ Blasiak, S.; Rangwala, H. (2011). "A Hidden Markov Model Variant for Sequence Classification". IJCAI Proceedings-International Joint Conference on Artificial Intelligence. 22: 1192.
- ^ Wong, W.; Stamp, M. (2006). "Hunting for metamorphic engines". Journal in Computer Virology. 2 (3): 211–229. doi:10.1007/s11416-006-0028-7. S2CID 8116065.
- ^ Wong, K. -C.; Chan, T. -M.; Peng, C.; Li, Y.; Zhang, Z. (2013). "DNA motif elucidation using belief propagation". Nucleic Acids Research. 41 (16): e153. doi:10.1093/nar/gkt574. PMC 3763557. PMID 23814189.
- ^ Shah, Shalin; Dubey, Abhishek K.; Reif, John (2019-05-17). "Improved Optical Multiplexing with Temporal DNA Barcodes". ACS Synthetic Biology. 8 (5): 1100–1111. doi:10.1021/acssynbio.9b00010. PMID 30951289. S2CID 96448257.
- ^ Shah, Shalin; Dubey, Abhishek K.; Reif, John (2019-04-10). "Programming Temporal DNA Barcodes for Single-Molecule Fingerprinting". Nano Letters. 19 (4): 2668–2673. Bibcode:2019NanoL..19.2668S. doi:10.1021/acs.nanolett.9b00590. ISSN 1530-6984. PMID 30896178. S2CID 84841635.
- ^ "ChromHMM: Chromatin state discovery and characterization". compbio.mit.edu. Retrieved 2018-08-01.
- ^ El Zarwi, Feraz (May 2011). "Modeling and Forecasting the Evolution of Preferences over Time: A Hidden Markov Model of Travel Behavior". arXiv:1707.09133 [stat.AP].
- ^ Morf, H. (Feb 1998). "The stochastic two-state solar irradiance model (STSIM)". Solar Energy. 62 (2): 101–112. Bibcode:1998SoEn...62..101M. doi:10.1016/S0038-092X(98)00004-8.
- ^ Munkhammar, J.; Widén, J. (Aug 2018). "A Markov-chain probability distribution mixture approach to the clear-sky index". Solar Energy. 170: 174–183. Bibcode:2018SoEn..170..174M. doi:10.1016/j.solener.2018.05.055. S2CID 125867684.
- ^ Munkhammar, J.; Widén, J. (Oct 2018). "An N-state Markov-chain mixture distribution model of the clear-sky index". Solar Energy. 173: 487–495. Bibcode:2018SoEn..173..487M. doi:10.1016/j.solener.2018.07.056. S2CID 125538244.
- ^ Baum, L. E.; Petrie, T. (1966). "Statistical Inference for Probabilistic Functions of Finite State Markov Chains". The Annals of Mathematical Statistics. 37 (6): 1554–1563. doi:10.1214/aoms/1177699147.
- ^ Baum, L. E.; Eagon, J. A. (1967). "An inequality with applications to statistical estimation for probabilistic functions of Markov processes and to a model for ecology". Bulletin of the American Mathematical Society. 73 (3): 360. doi:10.1090/S0002-9904-1967-11751-8. Zbl 0157.11101.
- ^ Baum, L. E.; Sell, G. R. (1968). "Growth transformations for functions on manifolds". Pacific Journal of Mathematics. 27 (2): 211–227. doi:10.2140/pjm.1968.27.211.
- ^ Baum, L. E.; Petrie, T.; Soules, G.; Weiss, N. (1970). "A Maximization Technique Occurring in the Statistical Analysis of Probabilistic Functions of Markov Chains". The Annals of Mathematical Statistics. 41 (1): 164–171. doi:10.1214/aoms/1177697196. JSTOR 2239727. MR 0287613. Zbl 0188.49603.
- ^ Baum, L.E. (1972). "An Inequality and Associated Maximization Technique in Statistical Estimation of Probabilistic Functions of a Markov Process". Inequalities. 3: 1–8.
- ^ Baker, J. (1975). "The DRAGON system—An overview". IEEE Transactions on Acoustics, Speech, and Signal Processing. 23: 24–29. doi:10.1109/TASSP.1975.1162650.
- ^ Jelinek, F.; Bahl, L.; Mercer, R. (1975). "Design of a linguistic statistical decoder for the recognition of continuous speech". IEEE Transactions on Information Theory. 21 (3): 250. doi:10.1109/TIT.1975.1055384.
- ^ Xuedong Huang; M. Jack; Y. Ariki (1990). Hidden Markov Models for Speech Recognition. Edinburgh University Press. ISBN 978-0-7486-0162-2.
- ^ Xuedong Huang; Alex Acero; Hsiao-Wuen Hon (2001). Spoken Language Processing. Prentice Hall. ISBN 978-0-13-022616-7.
- ^ Carrasco, Rafael C.; Oncina, Jose (1994). Carrasco, Rafael C.; Oncina, Jose (eds.). "Learning stochastic regular grammars by means of a state merging method". Grammatical Inference and Applications. Berlin, Heidelberg: Springer: 139–152. doi:10.1007/3-540-58473-0_144. ISBN 978-3-540-48985-6.
- ^ M. Bishop and E. Thompson (1986). "Maximum Likelihood Alignment of DNA Sequences". Journal of Molecular Biology. 190 (2): 159–165. doi:10.1016/0022-2836(86)90289-5. PMID 3641921. (subscription required)

- ^ Durbin, Richard M.; Eddy, Sean R.; Krogh, Anders; Mitchison, Graeme (1998), Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids (1st ed.), Cambridge, New York: Cambridge University Press, ISBN 0-521-62971-3, OCLC 593254083
- ^ Gassiat, E.; Cleynen, A.; Robin, S. (2016-01-01). "Inference in finite state space non parametric Hidden Markov Models and applications". Statistics and Computing. 26 (1): 61–71. doi:10.1007/s11222-014-9523-8. ISSN 1573-1375.
- ^ Abraham, Kweku; Gassiat, Elisabeth; Naulet, Zacharie (March 2023). "Fundamental Limits for Learning Hidden Markov Model Parameters". IEEE Transactions on Information Theory. 69 (3): 1777–1794. arXiv:2106.12936. doi:10.1109/TIT.2022.3213429. ISSN 0018-9448.
- ^ Beal, Matthew J., Zoubin Ghahramani, and Carl Edward Rasmussen. "The infinite hidden Markov model." Advances in neural information processing systems 14 (2002): 577-584.
- ^ Teh, Yee Whye, et al. "Hierarchical dirichlet processes." Journal of the American Statistical Association 101.476 (2006).
- ^ Ghahramani, Zoubin; Jordan, Michael I. (1997). "Factorial Hidden Markov Models". Machine Learning. 29 (2/3): 245–273. doi:10.1023/A:1007425814087.
- ^ Pieczynski, Wojciech (2002). "Chaı̂nes de Markov Triplet" (PDF). Comptes Rendus Mathématique. 335 (3): 275–278. doi:10.1016/S1631-073X(02)02462-7.
- ^ Pieczynski, Wojciech (2007). "Multisensor triplet Markov chains and theory of evidence". International Journal of Approximate Reasoning. 45: 1–16. doi:10.1016/j.ijar.2006.05.001.
- ^ Boudaren et al. Archived 2014-03-11 at the Wayback Machine, M. Y. Boudaren, E. Monfrini, W. Pieczynski, and A. Aissani, Dempster-Shafer fusion of multisensor signals in nonstationary Markovian context, EURASIP Journal on Advances in Signal Processing, No. 134, 2012.
- ^ Lanchantin et al., P. Lanchantin and W. Pieczynski, Unsupervised restoration of hidden non stationary Markov chain using evidential priors, IEEE Transactions on Signal Processing, Vol. 53, No. 8, pp. 3091-3098, 2005.
- ^ Boudaren et al., M. Y. Boudaren, E. Monfrini, and W. Pieczynski, Unsupervised segmentation of random discrete data hidden with switching noise distributions, IEEE Signal Processing Letters, Vol. 19, No. 10, pp. 619-622, October 2012.
- ^ Sotirios P. Chatzis, Dimitrios Kosmopoulos, "Visual Workflow Recognition Using a Variational Bayesian Treatment of Multistream Fused Hidden Markov Models," IEEE Transactions on Circuits and Systems for Video Technology, vol. 22, no. 7, pp. 1076-1086, July 2012.
- ^ Chatzis, Sotirios P.; Demiris, Yiannis (2012). "A Reservoir-Driven Non-Stationary Hidden Markov Model". Pattern Recognition. 45 (11): 3985–3996. Bibcode:2012PatRe..45.3985C. doi:10.1016/j.patcog.2012.04.018. hdl:10044/1/12611.
- ^ M. Lukosevicius, H. Jaeger (2009) Reservoir computing approaches to recurrent neural network training, Computer Science Review 3: 127–149.
- ^ Azeraf, E., Monfrini, E., & Pieczynski, W. (2023). Equivalence between LC-CRF and HMM, and Discriminative Computing of HMM-Based MPM and MAP. Algorithms, 16(3), 173.
- ^ Azeraf, E., Monfrini, E., Vignon, E., & Pieczynski, W. (2020). Hidden markov chains, entropic forward-backward, and part-of-speech tagging. arXiv preprint arXiv:2005.10629.
- ^ Azeraf, E., Monfrini, E., & Pieczynski, W. (2022). Deriving discriminative classifiers from generative models. arXiv preprint arXiv:2201.00844.
- ^ Ng, A., & Jordan, M. (2001). On discriminative vs. generative classifiers: A comparison of logistic regression and naive bayes. Advances in neural information processing systems, 14.
- ^ Wiggins, L. M. (1973). Panel Analysis: Latent Probability Models for Attitude and Behaviour Processes. Amsterdam: Elsevier.
- ^ Bartolucci, F.; Farcomeni, A.; Pennoni, F. (2013). Latent Markov models for longitudinal data. Boca Raton: Chapman and Hall/CRC. ISBN 978-14-3981-708-7.
- ^ Sofic Measures: Characterizations of Hidden Markov Chains by Linear Algebra, Formal Languages, and Symbolic Dynamics - Karl Petersen, Mathematics 210, Spring 2006, University of North Carolina at Chapel Hill
- ^ a b Boyle, Mike; Petersen, Karl (2010-01-13), Hidden Markov processes in the context of symbolic dynamics, arXiv:0907.1858
External links
[edit]Concepts
[edit]- Teif, V. B.; Rippe, K. (2010). "Statistical–mechanical lattice models for protein–DNA binding in chromatin". J. Phys.: Condens. Matter. 22 (41): 414105. arXiv:1004.5514. Bibcode:2010JPCM...22O4105T. doi:10.1088/0953-8984/22/41/414105. PMID 21386588. S2CID 103345.
- A Revealing Introduction to Hidden Markov Models by Mark Stamp, San Jose State University.
- Fitting HMM's with expectation-maximization – complete derivation
- A step-by-step tutorial on HMMs Archived 2017-08-13 at the Wayback Machine (University of Leeds)
- Hidden Markov Models (an exposition using basic mathematics)
- Hidden Markov Models (by Narada Warakagoda)
- Hidden Markov Models: Fundamentals and Applications Part 1, Part 2 (by V. Petrushin)
- Lecture on a Spreadsheet by Jason Eisner, Video and interactive spreadsheet


















































